Going back in genes

A new analysis of the genes common to bacteria and archaea offers strong evidence that the earliest cells on Earth lived deep in hydrothermal vents, explains William F Martin
The Biologist 64(2) p20-23
The last universal common ancestor (LUCA) is the name given to the most recent common ancestor of all existing life on Earth. It is the population of cells from roughly four billion years ago from which all life found on Earth today evolved.
We know LUCA must have had a DNA-based genetic code, because all descendant life forms do. But how did LUCA harness energy? The chemical reactions that help cells harness energy from their environments today seem almost as diverse as life itself.
We looked at that question using data from sequenced microbial genomes[1]. We concluded that LUCA probably lived from gases – H2, CO2, CO, N2 – in a setting that looked very much like a modern submarine hydrothermal vent.
There have been many investigations of LUCA using genomes, and the phylogenetic framework for investigating the presence of genes of LUCA has changed in the past few years (see Fig. 1, below).
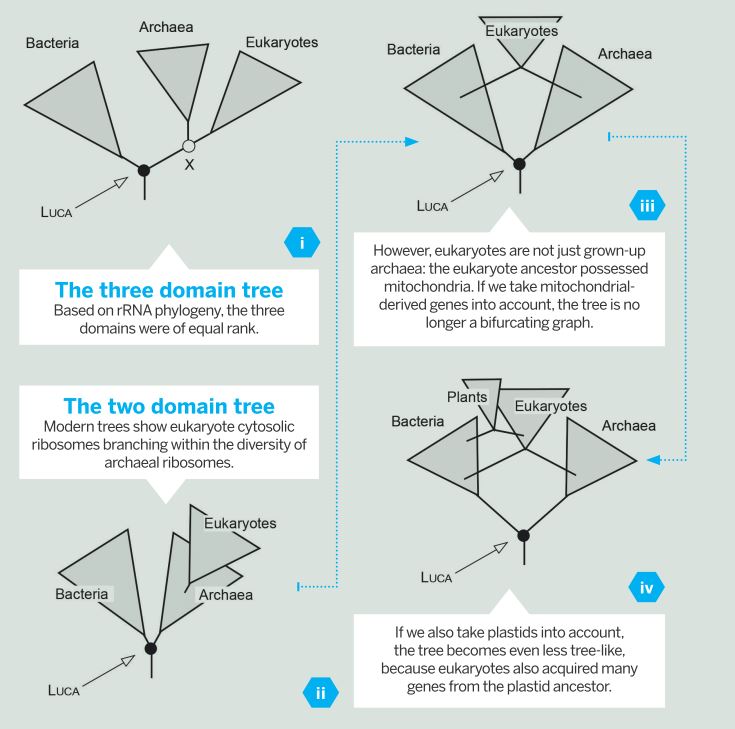 Fig. 1 (Click to enlarge) Many biologists are familiar with the three domain tree of life (i), which classifies all life forms that have ribosomes into archaea, bacteria, and eukaryotes. As more advanced phylogenetic data becomes available, new trees have emerged in which the eukaryotes branch within the archaeal diversification, not as sisters to them (ii).
Fig. 1 (Click to enlarge) Many biologists are familiar with the three domain tree of life (i), which classifies all life forms that have ribosomes into archaea, bacteria, and eukaryotes. As more advanced phylogenetic data becomes available, new trees have emerged in which the eukaryotes branch within the archaeal diversification, not as sisters to them (ii).The new 'two domain' tree of life affects many aspects of how we view microbial evolution – in particular, how we view LUCA. It means LUCA is the common ancestor of bacteria and archaea, with eukaryotes representing both bacterial (mitochondrial) and archaeal (host) genes at eukaryote origin (iii, iv). This means gene presence or absence in eukaryotes has no direct bearing on gene presence or absence in LUCA.
The classical approach is to look at a sample of genomes and see what is universally present in all of them. Genes that are present in all modern forms of life, by inference, are said to be present in LUCA, too (Fig. 2a, below).
Applying this strictly, around 35 genes trace to LUCA[2]. However, if this criteria is relaxed a bit to allow for a gene to have been lost in some lineages, the list grows to about 100 genes[3].
Lists of universally (or almost universally) conserved genes typically involve proteins associated with the ribosome (translation) and other aspects of information processing. These genes tell us that LUCA harnessed energy, as protein synthesis is the most expensive function of a cell (around 75% of a cell's ATP synthesis is spent on protein synthesis)[4]. However, these universal genes do not tell us what we wanted to know – namely, how the first cells harnessed energy.
A new approach to reconstructing LUCA genes from genomes is to allow not just a little bit of loss, but to allow loss quite freely. This way of investigating genome data has it that any gene that is present in bacteria and archaea – the two primordial domains of life – could also have been present in LUCA[5].
The problem with that approach, however, is that genes present in archaea and bacteria could have come to their present distribution in one of two ways.
They could have been present in LUCA and differentially lost. Or they could have evolved in a lineage that lived long after LUCA and subsequently passed from bacteria to archaea (or vice versa) via lateral gene transfer (LGT) – that is, the movement of genetic material between species of unicellular and/or multicellular organisms (Fig. 2b).
LGT can occur through many mechanisms, from the simple uptake of one prokaryote's genetic material by another to the transmission of DNA from one species to another via viruses.
Research has shown that thousands of such 'transdomain' LGTs have occurred during evolution[6]. Evolutionarily late, lineage-specific gene invention in bacteria followed by LGT across the domain divide separating bacteria from archaea mean genes can end up in both domains.
As such, we took a different approach to investigating LUCA with genome data. Among six million proteins encoded in 2,000 genomes, we looked not for the genes that are universal, and not for those that are just present in archaea and bacteria, but asked which genes are present in bacteria and archaea, and not as the result of LGT. Therefore, those genes should have been present in LUCA and vertically inherited within the domains since LUCA's time.
To do this, you must make phylogenetic trees – lots of them. Make trees of every gene that will produce a tree and filter them by criteria that will separate the vertical inheritance cases (the wheat) from the LGT cases (the chaff). Our method for distinguishing between vertical inheritance and LGT was straightforward.
First, the tree needed to contain sequences from both archaeal and bacterial genomes with a common root. Second, it needed to contain representatives from at least two archaeal phyla (a higher-order taxonomic group) and two bacterial phyla (familiar bacterial phyla being alphaproteobacteria, actinobacteria or cyanobacteria, for example). Although it is not impossible for LGT to produce a tree that fulfils both criteria, it is definitely not the null hypothesis.
Indeed, among 11,093 trees that contained sequences from both archaea and bacteria, only 355 trees satisfied both criteria. Of course, high-powered computers do all the nitty-gritty work, which still takes many months. When the results came in, though, our list contained 355 genes that were not ancient by virtue of simple distribution criteria – present in archaea and bacteria – they were ancient by phylogenetic criteria. That was new (see Fig. 2c).
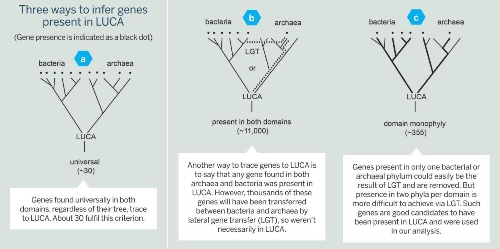 Fig 2 (click to enlarge) Showing three approaches to determining genes that may have been present in LUCA. Figure c) is the approach taken by the author, yielding 355 genes.
Fig 2 (click to enlarge) Showing three approaches to determining genes that may have been present in LUCA. Figure c) is the approach taken by the author, yielding 355 genes. The number of genes we found was not the surprise; the surprise was what the genes encoded. They encoded enzymes typical of modern cells that live in the strict absence of oxygen and in the presence of H2, CO2, CO, and N2, growth substrates that existed on the early Earth.
There were also clear hints in the data that LUCA was a thermophile. Furthermore, LUCA's enzymes were replete with transition metal electron carriers and catalysts – in particular, ferrous sulfide and ferrous-nickel sulfide centres. Taken together, that indicates that the last common ancestor of all cells grew from gases in a hot environment where metals and metal sulfides were abundant.
The core of LUCA's energy metabolism looked very similar to what we see in modern anaerobic, H2-dependent chemolithoautotrophs. In a nutshell, we found that LUCA had the genes for a lifestyle very similar to some modern prokaryotes – specifically, bacterial acetogens (acetate-producing anaerobes) and archaeal methanogens (methane-producing anaerobes).
Methanogens and acetogens inhabit a wide range of strictly anaerobic environments where H2, their main chemical fuel for CO2 reduction, is abundant. That can be the digestive tract of animals, which is not an ancient environment. It can be organic sediment at the bottom of lakes and oceans, also an environment dependent on other biomass, and therefore not an ancient environment. Or it can be the Earth's crust, which is an ancient environment. Both methanogens and acetogens are found today in the Earth's crust[8], where they harness energy by making methane and acetate out of H2, CO2 and CO.
Around four billion years ago, when LUCA lived[9], there was abundant CO2 – perhaps 1,000 times more than in the oceans today – but H2 was more restricted in terms of supply outlets. So where did the fuel for LUCA's metabolism come from? Today, there are two main sources of H2 in the environment. It is produced by microbes during the fermentation of decaying biomass and it is made geochemically in the Earth's crust via a process called serpentinisation, which occurs when water is circulated through the crust in hydrothermal systems.
LUCA was a pioneer on a previously uninhabited planet full of rocks, water and CO2, which meant biological H2 was not available. Geochemical H2 was abundantly available, though: geochemists tell us that hydrothermal vents have been producing H2 in abundance since there was water on Earth[10].
If the first forms of life survived from H2 and CO2, like LUCA's data suggest, then the first environment that was truly colonised by life was not the ocean, but the crust. That is an interesting thought when it comes to looking for life on other celestial bodies.
Interestingly, no traces of light utilisation turned up in LUCA's genome data – LUCA lived from chemical energy. For it to flourish, all that was required were rocks, metals, H2, CO2, water and hydrothermal activity. That finding means sunlight or ultraviolet light was not required to get LUCA going or to keep it alive. That, in turn, means that in the search for life elsewhere in our solar system, light need not be a limiting factor.
That has implications in particular for faraway moons such as Enceladus, which orbits Saturn and which has a liquid water ocean, a rocky, metal-rich core and hydrothermal activity, possibly involving serpentinisation[11] at its south pole[12]. Such a chemical environment could, in principle, support the emergence of an organism like our own universal common ancestor.
Whether future missions to Enceladus will detect evidence for the existence of any complex chemical reactions that might provide hints of something akin to life remains to be seen. The possibility is quite real that at least some interesting rock-water-carbon chemistry is going on out there in complete darkness, under the ice of a faraway moon.
William F Martin is a professor at the Institute of Molecular Evolution, University of Düsseldorf, Germany, leading research into chloroplasts, mitochondria and eukaryotes. He is currently the editor-in-chief of Genome Biology and Evolution.
1) Weiss, M. C. et al. The physiology and habitat of the last universal common ancestor. Nature Microbiol. 1, 16116 (2016).
2) Dagan, T. & Martin, W. The tree of one percent. Genome Biol. 7(118) (2006).
3) Puigbò, P. et al. Search for a 'Tree of Life' in the thicket of the phylogenetic forest. J. Biol. 8(59) (2009).
4) Stouthamer, H. Energy Yielding Pathways, in The Bacteria Volume 4 (eds Hrsg. Gunsalus, I. C. & Stanier, R. Y.) 389–462 (Academic Press, New York, 1978).
5) Baymann, F. et al. The redox protein construction kit: Pre-last universal common ancestor evolution of energy-conserving enzymes. Philos. Trans. R. Soc. Lond. 358, 267–274 (2003).
6) Nelson-Sathi, S. et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80 (2015).
7) Chapelle, F. H. et al. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415, 312–315 (2002).
8) Lever, M. A. et al. Acetogenesis in deep subseafloor sediments of the Juan de Fuca Ridge Flank. Geomicrobiol. J. 27, 183–211 (2010).
9) Schönheit, W. et al. On the origin of heterotrophy. Trends Microbiol. 24, 12–25 (2016).
10) Sleep, N. H. et al. Serpentinite and the dawn of life. Philos. Trans. R. Soc. Lond. B. 366, 2857–2869 (2011).
11) McCollom, T. M. Abiotic methane formation during experimental serpentinization of olivine. Proc. Natl. Acad. Sci. USA 113, 13965–13970 (2016).
12) Hsu, H. W. et al. Ongoing hydrothermal activities within Enceladus. Nature 519, 207–210 (2015).
13) Williams, T. A. et al. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236 (2013).
14) Cox, C. J. et al. The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. USA 105, 20356–20361 (2008).
15) Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
16) Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
17) Ku, C. et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432 (2015).


