Redefining Life
Rodrigo Rodrigues and Jônatas Abrahão look at so-called giant viruses – and how their unique features have forced us to reconsider whether viruses are living organisms or not
The Biologist 63(2) p12-15
Viruses were discovered towards the end of the 19th century, thanks to the pioneering studies of Louis Pasteur, and Dmitri Iwanowsky and Martinus Beijerinck's separate work on tobacco mosaic disease.
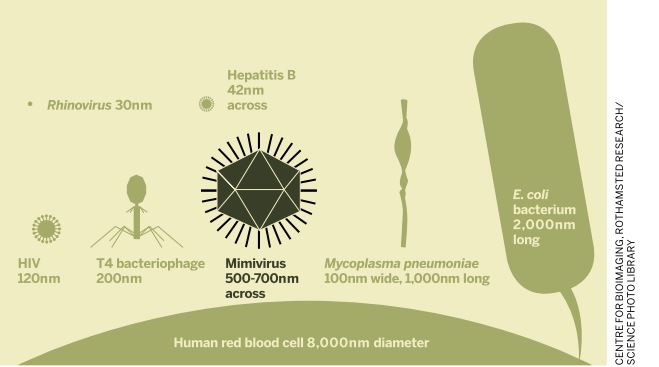
They were first defined as nanoscopic filterable agents with a maximum size of about 200nm.
In 1957, André Lwoff published a paper[1] on the basic features for an organism to be considered a virus: infectious; potentially pathogenic; but presenting only one kind of nucleic acid, DNA or RNA; unable to grow and replicate by binary fission, and lacking its own metabolic machinery.
Viruses have also traditionally been considered a group of polyphyletic microorganisms (having many origins) and kept out of the tree of life. Are all these definitions still valid to virologists and biologists today? A discovery in 2003 changed most of them.
The year was a milestone for virology. A new virus, called Acanthamoeba polyphaga mimivirus, was the first of many pathogens to be discovered that are now known as giant viruses.
The history of giant viruses began in 1992, in Bradford, England. Researchers were working on a pneumonia outbreak and in their search for pathogenic microorganisms they found an organism similar to a Gram-positive coccus, isolated in a culture of the amoeba Acanthamoeba polyphaga in a water sample from a hospital cooling tower. The pathogen, named Bradfordcoccus, was impossible to label with classical techniques for bacterial identification.
After several attempts to identify the new microorganism, the original sample was stored away until the early 2000s, when it was taken to the University of Marseille, France. There, a team led by Dr Bernard La Scola and Dr Didier Raoult resumed the investigation.
They decided to look directly at it using transmission electron microscopy. To the entire scientific community's surprise, Bradfordcoccus was actually an enormous virus, with very unusual structures never seen before in the virosphere[2].
It was renamed Acanthamoeba polyphaga mimivirus (APMV), a reference to the host in which it was first isolated and the fact it was able to mimic a microorganism. For its unusual features to be properly accommodated, a new viral family was created, named Mimiviridae. The APMV presented some features that suggested it was related to other large viruses, generically named as nucleo-cytoplasmic large DNA viruses (NCLDV, a proposed order named Megavirales).
This group comprises the families Poxviridae (including Variola virus), Iridoviridae (insect-infecting viruses) and Phycodnaviridae (algae-infecting viruses), among others, as well as some newly discovered but unclassified giant viruses, such as Pithovirus, Pandoravirus and Mollivirus.
After analysis of the genome of APMV, this hypothesis was confirmed, grouping it definitively with the other large viruses. In addition, the discovery of other mimiviruses in many parts of the world (North and South America, Europe and Africa so far), legitimised the existence of the Mimiviridae family.
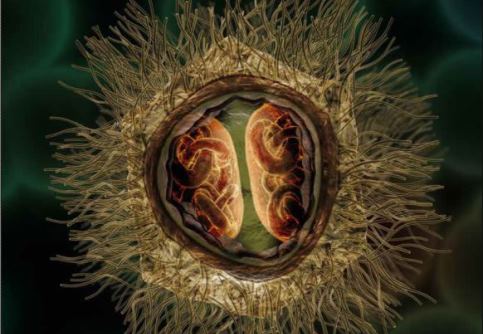
So what makes this virus so different? At about 750nm in diameter, it is far bigger than any other known viruses at the time of its discovery, and even bigger than some bacteria such as Mycoplasma and Rickettsia. Its capsid is pseudo-icosahedral (not the classic icosahedron), due to a star-shaped structure in one of the capsid vertex, named stargate, which is responsible for the genome release into the cytoplasm of a host cell.
Inside the capsid there is a lipid membrane, surrounding a proteic core that conceals the genome. Also, the capsid is covered by a dense layer of glycoproteic fibrils of approximately 125nm in length. The only region that lacks fibrils is the stargate, and one can easily see this in a scanning electron microscopy image. These fibrils are immersed in a peptidoglycan matrix, which is why the mimivirus was considered positive in the Gram staining.
Considering the virus's size and the presence of peptidoglycan, it is not surprising that those researchers in 1992 thought they were dealing with a bacterium.
Thanks to the fibrils, the mimiviruses are capable of attaching to several environmental organisms, such as amoebae, Gram-positive bacteria, fungi and arthropods[3].
Their ability to attach to arthropods such as crustaceans and mosquitoes might account for the ubiquity of these viruses, as these animals could disperse the giant viruses to many places on the planet, and also come into contact with humans.
One of the largest phage viruses, the T4 bacteriophage, is around half the size of the mimivirus and has a fraction of the number of base pairs (69,000). The mimivirus is around the same size as the Staphylococcus aureus bacterium.
The differences are not restricted to the viruses' structure - the genome of APMV is also distinct and complex. Its linear double-strand DNA molecule is 1.2 million base pairs – larger than the bacterium Mycoplasma pneumonia and most viruses, whose genomes are normally measured in thousands of base pairs.
The APMV genome's content is impressive too. Among its almost 1,000 genes, genes related to protein translation apparatus (normally associated only with cellular organisms) were found. There were also enzymes related to DNA repair (e.g. endonucleases), RNA modification (e.g. tRNA-methyltransferases) and carbohydrate metabolism (e.g. glycosyltransferases)[4]. Some transfer RNA (tRNA) was also identified within the viral particle.
The presence of classical cell genes in the mimivirus's genome dramatically changed scientists' view about viruses, reopening an old debate: are viruses living organisms?
Before the discovery of the mimiviruses, a consensus considered viruses to be 'just' small, obligate, intracellular microorganisms, completely dependent on cellular machinery, with few reasons to group them along with cellular or living organisms. However, the huge size and presence of classical cell genes within mimivirus's genome put this consensus to the test. For the first time, it was possible to phylogenetically cluster the viruses along with the other domains of life, Eukarya, Bacteria and Archaea in a metaphorical tree of life, leading to the proposal of a fourth domain of life, which includes the viruses from Megavirales[5].
Moreover, with genes related to translational apparatus, the mimiviruses possess a partial independence from the host. Since they can also be parasitised by other viruses (virophages), it reinforced the idea that viruses could be regarded as living organisms, and so should be represented in the tree of life. However, in science, new proposals are not accepted easily.
Some scientists are against the creation of a fourth domain of life for a number of reasons. Even though the mimiviruses have characteristics that partially exclude them from the classical definition of viruses proposed by André Lwoff, and have a relative independence from the host, they are still inert out of a cell. In addition, the viruses do not have a single ancestral lineage. They are polyphyletic organisms, and some of their genes come from horizontal gene transfer; it is impossible to establish a reliable phylogenetic relationship based on these genes[6]. Others believe what should be considered is the virus while inside the cell host[7] ('the virocell'), which forms a viral factory and is able to replicate and evolve, both essential characteristics of life.
The debate will rumble on, but the entire classification system could change in the coming years. It is already feasible that all the organisms on the planet will be divided into two groups, ribosome encoding organisms (i.e. cellular organisms, or REOs) and capsid encoding organisms (viral organisms, or CEOs), leaving the idea of domains behind. Some evolutionists still claim that the term 'domain' should not be extinguished, but used exclusively for REO, and the term 'lineage' should be for CEO members. Only time will tell if these ideas succeed.
There is still a lot to discover. Whether viruses will be considered alive or not is uncertain, but no one can deny the role they play in the evolution of life.
Jônatas Abrahão is professor of virology at Federal University of Minas Gerais, Brazil. His research interests focus on relationships among large/giant viruses and their hosts.
Rodrigo Rodrigues is a PhD student in the Department of Microbiology at the Federal University of Minas Gerais, Brazil. His research interests focus on the structural, ecological and evolutionary aspects of giant viruses.
Are giant viruses a threat to people?
It has been shown that mimiviruses can enter and replicate in human macrophages and in certain blood cells, where they interfere with the innate immune system[8]. There is evidence that they infect humans (such as the detection of mimiviruses and anti-mimivirus antibodies in patients), but there is still no consensus[9,10].
It remains a mystery whether they caused the pneumonia outbreak that occurred when they were first isolated in Bradford in 1992.
1) Lwoff, A. The concept of virus. J Gen Microbiol 17(2), 239–253 (1957).
2) La Scola, B. et al. A giant virus in amoebae. Science 299(5615), 2033 (2003).
3) Rodrigues, R. A. et al. Mimivirus fibrils are important for viral attachment to the microbial world by a diverse glycoside interaction repertoire. J Virol 89(23), 11812–11819 (2015).
4) Raoult, D. et al. The 1.2-megabase genome sequence of Mimivirus. Science 306(5700), 1344–1350 (2004).
5) Boyer, M. et al. Phylogenetic and phyletic studies of informational genes in genome highlight existence of a 4th domain of life including giant viruses. PLoS One 5(12), e15530 (2010).
6) Moreira, D. & López-García, P. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7(4), 306–311 (2009).
7) Claverie, J. M. & Ogata, H. Ten good reasons not to exclude giruses from the evolutionary picture. Nat Rev Microbiol 7(8), 615 (2009).
8) Silva, L. C. et al. A resourceful giant: APMV is able to interfere with the human type I interferon system. Microbes and Infect 16(3), 187–195 (2014).
9) La Scola, B. et al. Mimivirus in pneumonia patients. Emerg Infect Dis 11(3), 449–452 (2005).
10) Saadi, H. et al. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis 54(7), e127–134 (2013).