Hunting for 'holes of hope'
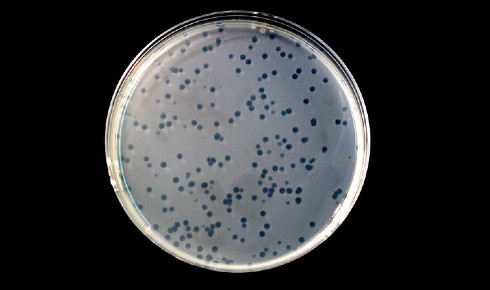
Above: the characteristic holes or 'plaques' indicating bacteriophage infection of a plate of bacteria. Courtesy of Ninjatacoshell via Wikimedia commons.
Bacteriophages found by members of the public are helping create libraries of viruses that could be used to treat multidrug resistant infections, writes Dr Ben Temperton
October 25th 2021
It is perhaps comforting to think that pandemics are rare, and that by the next one, we will have developed more effective and equitable ways of responding. In reality, the next global health pandemic is already here, in the form of antimicrobial resistance. Like COVID-19, the international movement of goods and people has spread antibiotic resistant bacteria across the globe, but the threat we face from antimicrobial resistance is not from one type of pathogen, but hundreds.
In 2018, approximately 28% of bacterial infections were found to be resistant to antibiotics. By 2050 it is estimated this will increase to 40% and result in an additional 10 million deaths a year.
The pipeline for new antibiotics has run dry because the economics of developing new antibiotics for profit do not add up. The for-profit model of drug development by private enterprise has so-far proven poorly equipped to solve the resistance pandemic. If we are to meet this new threat, it is time for a different approach.
Let us imagine for a moment what an ideal new class of antimicrobials would look like. They would be cheap to produce, with minimal equipment. They would be easily stored, shipped and administered. They would be readily adaptable to meet emerging bacterial threats as new resistances emerged. They would work in synergy with existing antibiotics, extending the utility of what we already have.
Unlike current last-line-of-defence antibiotics, they would be free from harmful side effects and highly specific, killing the pathogen but leaving the remainder of the microbiome intact to avoid issues of chronic remission observed in other long term antibiotic treatments. To ensure equitable global access and availability, they would be unprotected by patent, produced and distributed under a commons licence as public goods.
There is no doubt that a novel class of drug that met these criteria would be hailed as a major medical advance. It is then perhaps surprising that antimicrobials with these properties already exist and occur naturally in the environment. Not only have we known about them for over 100 years, they have been used to treat bacterial infections since their discovery.
 Phages bind to receptors on their bacterial prey using protein tail fibres (the leg-like structures on the illustration above) before injecting their genetic material into the host cell. If the phage genes can evade the bacterium's defences, the cell effectively becomes a phage factory that can produce hundreds of new progeny. Once assembly is complete, the cell is destroyed, releasing the new phages into the environment to seek out new hosts.
Phages bind to receptors on their bacterial prey using protein tail fibres (the leg-like structures on the illustration above) before injecting their genetic material into the host cell. If the phage genes can evade the bacterium's defences, the cell effectively becomes a phage factory that can produce hundreds of new progeny. Once assembly is complete, the cell is destroyed, releasing the new phages into the environment to seek out new hosts.Holes of hope
In 1916, before the discovery of the earliest antibiotics, Felix D’Herelle, an irascible French Canadian microbiologist at the Pasteur Institute in Paris, noticed clear holes (called ‘plaques’) in the lawns of bacteria he was culturing from cavalry troops suffering from dysentry caused by the bacteria Shigella. D’Herelle had discovered bacteriophages – the nanoscopic viruses that infect and kill bacteria.
Bacteriophages, or ‘phages’ as they are more commonly known, were independently discovered by an English doctor called Frederick Twort around the same time, but it was D’Herelle that coined the name (meaning ‘bacteria eater’) and pioneered their medical use.
D’Herelle’s work identified that phages were a clinically safe means of killing bacterial infections that were highly specific to different pathogenic strains. Immediately after their discovery, they were used to successfully treat (among other diseases) bubonic plague, cholera, dysentery and typhoid. In 1923 D’Herelle helped establish the Eliava Phage Therapy Centre in Tbilisi, Georgia that still treats patients to this day. At a time where a cut becoming infected could cause life-threatening illness and even minor surgery was a huge risk, interest and excitement of a safe way of treating bacterial infection grew.
Ultimately, however, it was the specificity of phage therapy and the need to isolate the infecting pathogen that saw its demise in the West when antibiotics like penicillin became widely available. These new pharmaceuticals were indiscriminate and could be given to patients if an infection was suspected, or even prophylactically. They could be manufactured at scale and patented.
The specificity of phages also hindered early clinical trials, where generic cocktails were tested against infectious bacteria that the phages couldn’t kill. Phage therapy fell out of favour in the West, but continued to be developed in Eastern countries where supply of antibiotics were limited.
Fifty years later, faced with the chastising threat of a post-antibiotic era, scientific interest in phage therapy is undergoing a global renaissance in the West. A lodestone for the renewed interest was the case of American academic Tom Patterson, who was struck down with an infection of Acinetobacter baumannii that was resistant to all known antibiotic treatments.
After nine months in hospital and multiple bouts of life-threatening sepsis, it was a combination therapy of antibiotics and phages that saved him. In the UK, Dr Helen Spencer used genetically engineered phages to cure Isabelle Carnell-Holdaway, a 15-year-old cystic fibrosis patient of multi drug-resistant Mycobacterium abscesses following a double lung transplant.
These exemplary stories, where experimental phage therapy was successfully used on compassionate grounds to treat the untreatable, shine a light on the growing number of uses of phage therapy across the globe, with over 600 applications of compassionate phage therapy since 2000.
Phage biobanks for clinical use are being developed or expanded in Australia, France, USA, South Korea, Israel, Belgium, Poland, and Georgia among others. Belgium has gone one step further and developed legal and clinical frameworks for phage therapy beyond compassionate use, with phages now available in combination with antibiotics through magisterial preparations within hospital pharmacies.
Phage hunters
 The Citizen Phage Library project encourages members of the public to find and name their own phage from water samples taken in their local area.
The Citizen Phage Library project encourages members of the public to find and name their own phage from water samples taken in their local area. Here at Exeter, the Citizen Phage Library is a new enterprise established to accelerate development of phage therapy. The project has several aims. Firstly, to rapidly develop a phage library in the UK for clinical use, targeting environmental phages from as broad a range of locations as possible to maximise diversity. We want to enable global sharing of fully characterised phages at cost to the clinicians who need them, and to accelerate phage research by supplying phage researchers with well-characterised phages for their studies.
We also hope that by engaging members of the public with our search for phages we can increase awareness of clinical phage therapy and antimicrobial resistance. A final aim is to provide an open-source, low-cost template for establishing a national phage library as a social enterprise that can be replicated in countries where resources are limited.
Citizen scientists are recruited at science fairs and school visits as “phage hunters”. Each phage hunter is provided with a sampling kit containing three 40ml jars and a pamphlet describing the project and its aims.
Phage hunters collect samples from water sources such as ponds, bird baths, rivers and streams and log them into the Citizen Phage Library website. Each sample is associated with a description of where it came from, including the geolocation of the collection point to direct resampling if interesting phages are found. This data will also support optimisation of future sampling campaigns when new phages are needed to treat a patient.
Samples are posted back to our lab, where they are screened against a panel of drug-resistant bacteria which pose the greatest risks to human health. First, the samples are centrifuged and filtered to remove any bacteria. They are then mixed with a culture of the target pathogen and incubated overnight at 37 °C. Any suitable phages found in the sample will attach to their host and kill it, producing more phages. Again, the bacteria is removed and the now enriched phage mixture is spotted onto a lawn of pathogenic bacteria to see if a ‘plaque’ appears to indicate that a phage is infecting and replicating on the lawn.
Each phage identified in a sample is purified from the plaques, imaged and their genomes are sequenced to ensure that phages do not contain any genes of concern, such as those encoding toxic metabolites, antibiotic resistance or anything that might actually increase the fitness of their bacterial hosts. Phages are also screened for synergistic interactions with antibiotics typically used in primary care interventions.
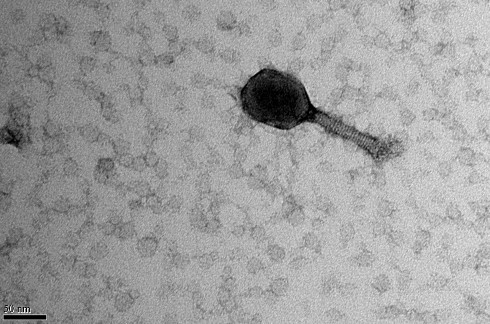 The bacteriophage 'LemonAid', discovered as part of the project in water from Devon's River Lemon, was found to infect and kill Acinetobacter baumannii, number one on the World Health Organisation's list of most concerning drug-resistant pathogens. (Image courtesy of Christian Hacker at Exeter University Bioimaging Centre)
The bacteriophage 'LemonAid', discovered as part of the project in water from Devon's River Lemon, was found to infect and kill Acinetobacter baumannii, number one on the World Health Organisation's list of most concerning drug-resistant pathogens. (Image courtesy of Christian Hacker at Exeter University Bioimaging Centre)The purified phage cultures are stored in our phage biobank and data describing the genome, antibiotic synergy and host range is stored in the Citizen Phage Library database. Once a phage has been characterised, the phage hunter is notified that a phage was found in their sample and they are asked if they would like to name the phage and be recorded in the database as its discoverer. Soon, this database will be integrated with the PhageDirectory service which connects global clinicians requiring phages with phage biobanks.
In the first few months of operation, the Citizen Phage Library has isolated 78 new phages from around 100 samples. Allowing citizens to name their phages and providing them with an electron micrograph of their discovery is met with great excitement and a source of wonderful creativity.
Examples of phages isolated from these citizen samples include phage “LemonAid”, isolated from the River Lemon in Devon, and a potent killer of a carbapenem-resistant Acinetobacter baumannii – classified by the World Health Organisation as a critical priority pathogen for new antimicrobials.
Phage “KylieMinegg” was recently isolated from a chicken coop and is a novel phage that efficiently kills a broad collection of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients.
Critically, isolating and characterising each phage is low-cost (a few £100) and takes 2-3 weeks, in contrast to the enormous time and financial costs of developing new antimicrobial drugs.
Portable sequencing technology and open source liquid-handling platforms minimise the need for expensive infrastructure. It is our hope that within five years, other national Citizen Phage Libraries will use our template to establish replicate efforts, each housing their own domestic collection of phages, with all associated data shared through a centralised database.
Phage therapy 2.0
Unlike the phage therapy of the 1920s and 30s, modern phage therapy is underpinned by rapid and cheap genome sequencing, high-resolution nanoscale imaging and molecular tools that can identify how phages interact with antibiotics and the immune system at the subcellular level.
The clinical safety of phages, particularly compared to some antibiotics, is enviable. Phages should be considered as safe, self-replicating biological agents that target a bacterial infection and leave the rest of the microbiome unaffected. The mechanisms of random mutation and selection that drive the evolution of resistance in bacteria to antibiotics are equally available to the phages, allowing them to overcome host resistance.
Recent research shows that treatment with both phages and antibiotics in unison (known as adjunctive therapy) can re-sensitise bacteria to previously ineffective antibiotics. It appears that bacteria can develop resistance to antibiotics, or even phages, but the fitness cost of evolving resistance to both simultaneously is too much to bear.
By swapping out the genes encoding the tail fibres of one phage with those from another, the host of a phage can be altered in a modular fashion. Modern machine-learning algorithms, trained on genomic information from phages and hosts, are being developed to optimise the creation of mixtures of phages and antibiotics for improved treatments.
Beyond direct use of natural phages in clinical treatments, the vast genetic diversity encoded within phage genomes is being tapped for development of novel, targeted therapeutics. There is little doubt that Phage Therapy 2.0, as it has been coined, offers a promising set of tools to meet the challenge of antimicrobial resistance.
Delivery on that promise is currently impeded by a lack of awareness among the public, clinicians and politicians of phage therapy. Furthermore, the use of personalised, biological agents such as phages in clinical treatment is far removed from prescribing chemical drugs for which a generalised response has been measured across a large group of people through clinical trials. Here, we can learn from our international colleagues to develop guidance for safe and effective use for compassionate phage therapy. Alternatives to large clinical trials, such as the ‘n-of-1’ trials being developed for personalised cancer treatment, should provide reassurance to clinicians for therapies beyond compassionate use. These barriers can be removed through provision of information and educational outreach to the public and clinicians alike.
Blockchain ledger technology can eventually facilitate rapid and seamless global sharing of phages, just as they are used to automate bills of lading for goods transport. Furthermore, the ledger will record the provenance of each phage in perpetuity, ensuring that the efforts of phage hunters and national biobanks are fully recognised. This ledger can be further expanded as a ‘living record’ for each phage, capturing details of each time it was used in phage therapy, such as treatment regime, which other phages and antibiotics it was used with and genomic information of the infecting pathogen. Such a public resource would be of huge benefit to train algorithms for optimising phage cocktails in future treatments.
As world leaders gather for the 26th United Nations Climate Change Conference, it is worth reflecting that the threat of antimicrobial resistance shares many features with the threat of climate change. There is near-universal consensus that both represent a major threat to global health, with the Global South predicted to bear the greatest cost.
It is also widely recognised that neither threat can be met by relying on for-profit companies to develop solutions when the incentives to do so are absent. Instead, a global coordinated response, supported by governments and driven by citizens is needed. Our vision for the Citizen Phage Library is to provide a platform that delivers the promise of Phage Therapy 2.0 globally, through the benefit of the commons. A distributed global network of social enterprises, funded by governments and philanthropy and linked through technology will ensure that solutions to the threat of antimicrobial resistance are available to all.
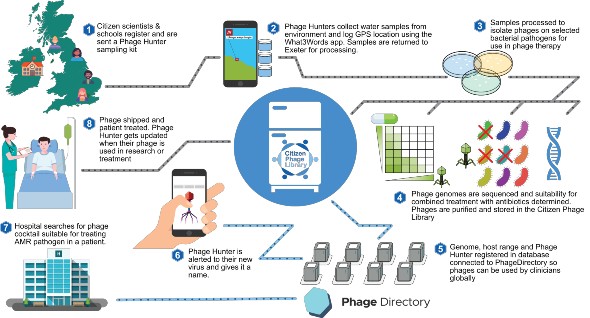 An overview of how citizen scientists can contribute to global biobanks of viruses for clinical use. Click to enlarge.
An overview of how citizen scientists can contribute to global biobanks of viruses for clinical use. Click to enlarge. Find out more about finding phages at www.citizenphage.com
Dr Ben Temperton is a senior lecturer in Bioinformatics at the University of Exeter and a microbial ecologist who researches how the interactions between bacteria and phages influence global biogeochemical cycles, and how we might be able to exploit bacteriophages to treat antibiotic resistant infections


